Using nrseq-fasta-importer¶
Overview¶
The nrseq-fasta-importer web application provides a dynamic GUI to parsing FASTA files and importing them into the YRC_NRSEQ database. For more information about what this means, see About nrseq-fasta-importer.
The following screenshot shows the interface:
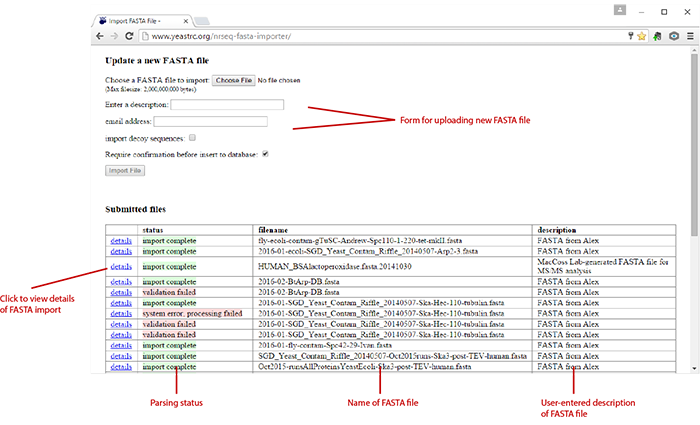
Upload the FASTA file¶
To upload a FASTA file, fill out the form at the top of the page. First click the
Choose File button and select the FASTA file. Enter a description for the
FASTA file, your email address (for status notifications), whether or not to
import decoy sequences, and whether or not to require user confirmation before
data is inserted into the YRC_NRSEQ database (recommended).
Once the form is submitted, an overlay will appear on the page:
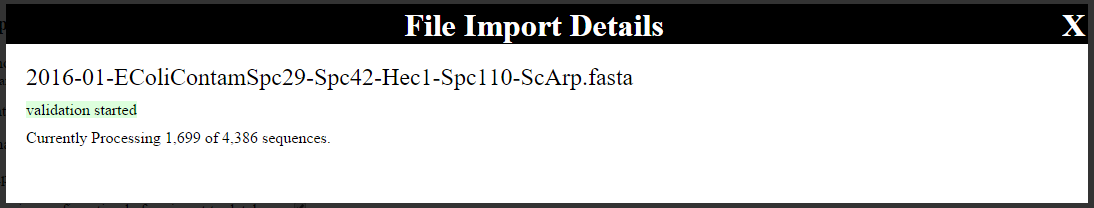
This overlay will display what is happening (FASTA validation, taxonomy determination, etc), and given a progress indication by showing how many sequences are left to process.
If validation fails, the overlay will indicate the failure. The FASTA file will need to be corrected and re-uploaded.
If taxonomy determine fails for too many entries (currently set to 200), the upload process will fail. Please see About nrseq-fasta-importer for more information about what to do in this situation.
User input of taxonomy IDs¶
If the taxonomy ID could not be determined for some of the FASTA entries (up to 200), an overlay will appear that allows users to manually enter the NCBI taxonomy ID for the FASTA headers:
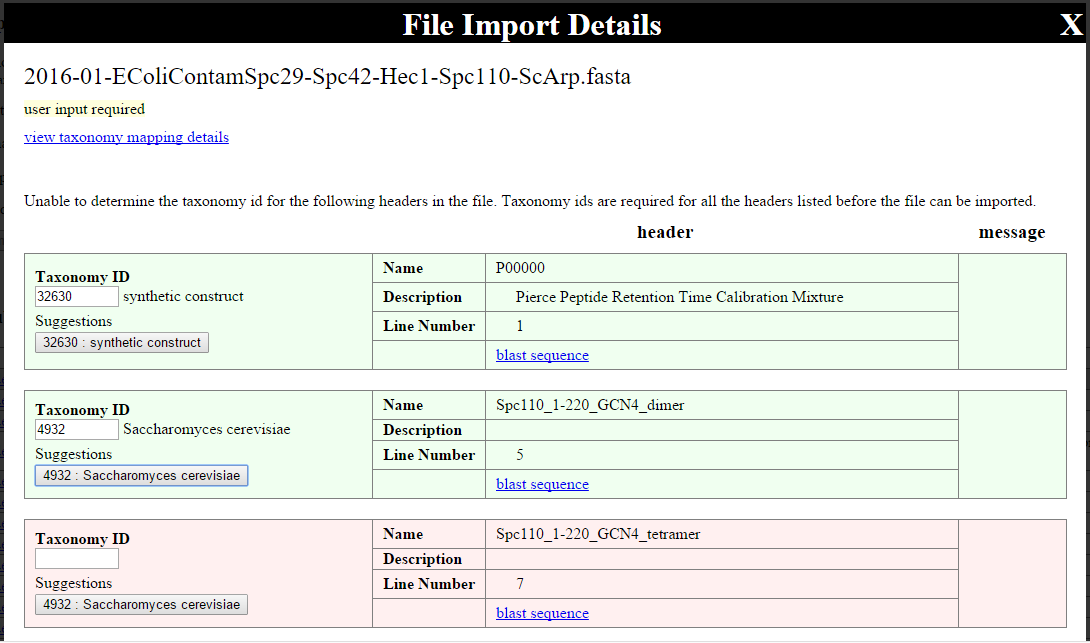
Each entry will be shaded red until a taxonomy ID is added, when it will turn green. When a taxonomy ID is entered, a web services lookup is made to NCBI to retrieve the name for the taxonomy, and this is displayed so that the user may verify the correctness of the ID.Suggestions for taxonomy ID are provided, based on taxonomy IDs associated with that sequence and name in the past. To choose the suggested taxonomy ID, click the button showing the suggestion.
Once all rows are green, click the button at the bottom of the list to re-initiate taxonomy determinations. Once this succeeds, the FASTA may be imported.
Inspect taxonomy ID assignments¶
To view the taxonomy ID assignments for the proteins in your FASTA file, click the
view taxonomy mapping details link present in the details overlay for the
FASTA file:
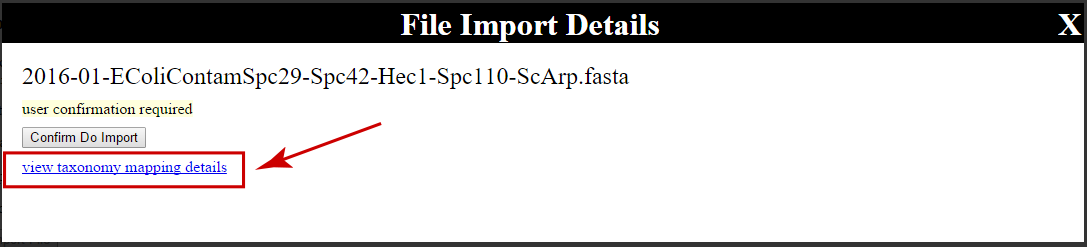
This will prompt the user to save an XML file, which may be viewed in a web browser or text editor. The file has the following syntax:
<intermediate-import-file>
<import-file-entry>
<headerLineNumber>1</headerLineNumber>
<importFileHeaderEntryList>
<headerDescription>Pierce Peptide Retention Time Calibration Mixture</headerDescription>
<headerName>P00000</headerName>
<taxonomyId>32630</taxonomyId>
</importFileHeaderEntryList>
<sequence>
SSAAPPPPPRGISNEGQNASIKHVLTSIGEKDIPVPKPKIGDYAGIKTASEFDSAIAQDKSAAGAFGPELSRELGQSGVDTYLQTKGLILVGGYGTRGILFVGSGVSGGEEGARSFANQPLEVVYSKLTILEELRNGFILDGFPRELASGLSFPVGFKLSSEAPALFQFDLK
</sequence>
</import-file-entry>
<import-file-entry>
<headerLineNumber>5</headerLineNumber>
<importFileHeaderEntryList>
<headerName>Spc110_1-220_GCN4_dimer</headerName>
<taxonomyId>4932</taxonomyId>
</importFileHeaderEntryList>
<sequence>
GSMDEASHLPNGSLKNMEFTPVGFIKSKRNTTQTQVVSPTKVPNANNGDENEGPVKKRQRRSIDDTIDSTRLFSEASQFDDSFPEIKANIPPSPRSGNVDKSRKRNLIDDLKKDVPMSQPLKEQEVREHQMKKERFDRALESKLLGKRHITYANSDISNKELYINEIKSLKHEIKELRKEKNDTLNNYDTLEEETDDLKNRLQALEKELDAKNKIVNSRKVDRMKQLEDKVEELLSKNYHLENEVARLKKLVGER
</sequence>
</import-file-entry>
<import-file-entry>
<headerLineNumber>7</headerLineNumber>
<importFileHeaderEntryList>
<headerName>Spc110_1-220_GCN4_tetramer</headerName>
<taxonomyId>4932</taxonomyId>
</importFileHeaderEntryList>
<sequence>
GSMDEASHLPNGSLKNMEFTPVGFIKSKRNTTQTQVVSPTKVPNANNGDENEGPVKKRQRRSIDDTIDSTRLFSEASQFDDSFPEIKANIPPSPRSGNVDKSRKRNLIDDLKKDVPMSQPLKEQEVREHQMKKERFDRALESKLLGKRHITYANSDISNKELYINEIKSLKHEIKELRKEKNDTLNNYDTLEEETDDLKNRLQALEKELDAKNKIVNSRKVDRMKQIEDKLEEILSKLYHIENELARIKKLLGER
</sequence>
</import-file-entry>
</intermediate-import-file>
There is a <import-file-entry> for each FASTA entry in the file. Each one contains the associated sequence, header(s) and associated taxonomy ID(s) found for that FASTA entry.
Import the FASTA file¶
If the Require confirmation before insert to database: option was not selected on the
upload form, import will automatically begin after successful validation and determination
of taxonomy IDs.
Otherwise, user confirmation is required. Confirmation may be given in the overlay showing the import status:

The status overlay for an import may be accessed by clicking the details link for the row
for that FASTA file in the interface or from the link in the status email received from
the web application.
Click Confirm Do Import to import the FASTA file to the database.
Upon successful completion, that status message will change to import complete and another
confirmation email will be sent.